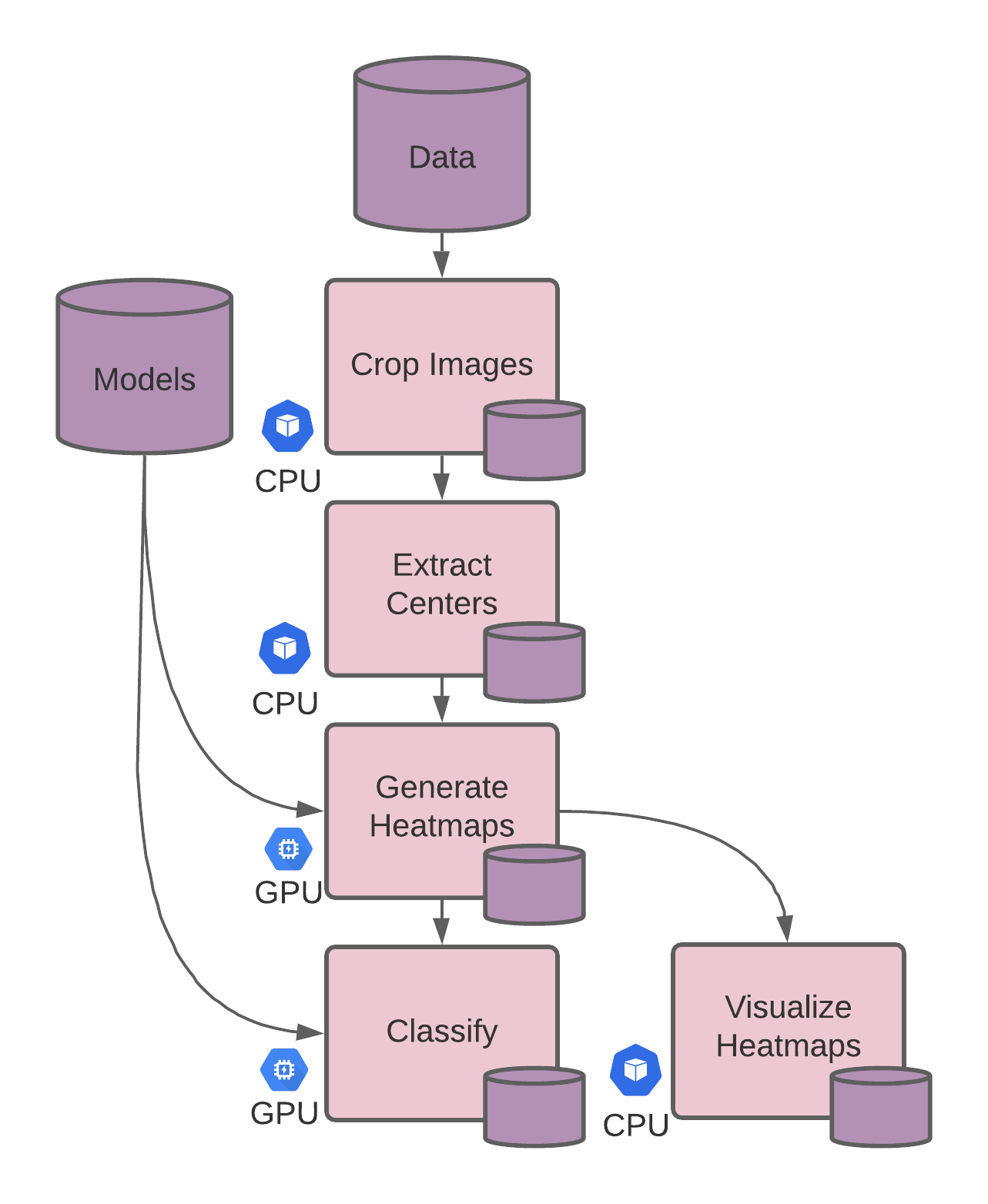
In this tutorial, we’ll build a scalable inference pipeline for breast cancer detection using task parallelism .
Before You Start
# Tutorial
# Our Docker image’s user code for this tutorial is built on top of the pytorch/pytorch base image, which includes necessary dependencies. The underlying code and pre-trained breast cancer detection model comes from this repo , developed by the Center of Data Science and Department of Radiology at NYU. Their original paper can be found here .
Pachctl CLI
Console
Make sure your Tutorials project we created in the Standard ML Pipeline tutorial is set to your active context. (This would only change if you have updated your active context since completing the first tutorial.)
pachctl config get context localhost : 80
# {
# "pachd_address": "grpc://localhost:80",
# "cluster_deployment_id": "KhpCZx7c8prdB268SnmXjELG27JDCaji",
# "project": "Tutorials"
# } Copy
Create the following repos:
pachctl create repo models
pachctl create repo sample_data Copy
Navigate to Console.
Select the previous standard-ml-pipeline project.
Create a models and sample_data repo.
2. Create CPU Pipelines
# In task parallelism, we separate out the CPU-based preprocessing and GPU-related tasks, saving us cloud costs when scaling. By separating inference into multiple tasks, each task pipeline can be updated independently, allowing ease of model deployment and collaboration.
We can split the run.sh script used in the previous tutorial (Data Parallelism Pipeline ) into 5 separate processing steps (4 already defined in the script + a visualization step) which will become Pachyderm pipelines, so each can be scaled separately.
Crop Pipeline
#
Pachctl CLI
Console
Create a file named crop.json with the following contents:
{
"pipeline" : {
"name" : "crop" ,
"project" : {
"name" : "standard-ml-pipeline"
},
},
"description" : "Remove background of image and save cropped files." ,
"input" : {
"pfs" : {
"repo" : "sample_data" ,
"glob" : "/*"
}
},
"transform" : {
"cmd" : [
"/bin/bash" ,
"multi-stage/crop.sh"
] ,
"image" : "pachyderm/breast_cancer_classifier:1.11.6"
}
} Copy
Save the file.
Create the pipeline.
pachctl create pipeline - f / path / to / crop.json Copy
Select Create > Pipeline .
Overwrite the default json with the following:
{
"pipeline" : {
"name" : "crop" ,
"project" : {
"name" : "standard-ml-pipeline"
},
},
"description" : "Remove background of image and save cropped files." ,
"input" : {
"pfs" : {
"repo" : "sample_data" ,
"glob" : "/*"
}
},
"transform" : {
"cmd" : [
"/bin/bash" ,
"multi-stage/crop.sh"
] ,
"image" : "pachyderm/breast_cancer_classifier:1.11.6"
}
} Copy
Pachctl CLI
Console
Create a file named extract_centers.json with the following contents:
{
"pipeline" : {
"name" : "extract_centers" ,
"project" : {
"name" : "standard-ml-pipeline"
},
},
"description" : "Compute and Extract Optimal Image Centers." ,
"input" : {
"pfs" : {
"repo" : "crop" ,
"glob" : "/*"
}
},
"transform" : {
"cmd" : [
"/bin/bash" ,
"multi-stage/extract_centers.sh"
] ,
"image" : "pachyderm/breast_cancer_classifier:1.11.6"
}
} Copy
Save the file.
Create the pipeline.
pachctl create pipeline - f / path / to / extract_centers.json Copy
Select Create > Pipeline .
Overwrite the default json with the following:
{
"pipeline" : {
"name" : "extract_centers" ,
"project" : {
"name" : "standard-ml-pipeline"
},
},
"description" : "Compute and Extract Optimal Image Centers." ,
"input" : {
"pfs" : {
"repo" : "crop" ,
"glob" : "/*"
}
},
"transform" : {
"cmd" : [
"/bin/bash" ,
"multi-stage/extract_centers.sh"
] ,
"image" : "pachyderm/breast_cancer_classifier:1.11.6"
}
} Copy
3. Create GPU Pipelines
# Generate Heatmaps Pipeline
#
Pachctl CLI
Console
Create a file named generate_heatmaps.json with the following contents:
{
"pipeline" : {
"name" : "generate_heatmaps" ,
"project" : {
"name" : "standard-ml-pipeline"
},
},
"description" : "Generates benign and malignant heatmaps for cropped images using patch classifier." ,
"input" : {
"cross" : [
{
"join" : [
{
"pfs" : {
"repo" : "crop" ,
"glob" : "/(*)" ,
"joinOn" : "$1" ,
"lazy" : false
}
},
{
"pfs" : {
"repo" : "extract_centers" ,
"glob" : "/(*)" ,
"joinOn" : "$1" ,
"lazy" : false
}
}
]
},
{
"pfs" : {
"repo" : "models" ,
"glob" : "/" ,
"lazy" : false
}
}
]
},
"transform" : {
"cmd" : [
"/bin/bash" ,
"multi-stage/generate_heatmaps.sh"
] ,
"image" : "pachyderm/breast_cancer_classifier:1.11.6"
},
"resourceLimits" : {
"gpu" : {
"type" : "nvidia.com/gpu" ,
"number" : 1
}
},
"resourceRequests" : {
"memory" : "4G" ,
"cpu" : 1
}
} Copy
Save the file.
Create the pipeline.
pachctl create pipeline - f / path / to / generate_heatmaps.json Copy
Select Create > Pipeline .
Overwrite the default json with the following:
{
"pipeline" : {
"name" : "generate_heatmaps" ,
"project" : {
"name" : "standard-ml-pipeline"
},
},
"description" : "Generates benign and malignant heatmaps for cropped images using patch classifier." ,
"input" : {
"cross" : [
{
"join" : [
{
"pfs" : {
"repo" : "crop" ,
"glob" : "/(*)" ,
"joinOn" : "$1" ,
"lazy" : false
}
},
{
"pfs" : {
"repo" : "extract_centers" ,
"glob" : "/(*)" ,
"joinOn" : "$1" ,
"lazy" : false
}
}
]
},
{
"pfs" : {
"repo" : "models" ,
"glob" : "/" ,
"lazy" : false
}
}
]
},
"transform" : {
"cmd" : [
"/bin/bash" ,
"multi-stage/generate_heatmaps.sh"
] ,
"image" : "pachyderm/breast_cancer_classifier:1.11.6"
},
"resourceLimits" : {
"gpu" : {
"type" : "nvidia.com/gpu" ,
"number" : 1
}
},
"resourceRequests" : {
"memory" : "4G" ,
"cpu" : 1
}
} Copy
Classify Pipeline
#
Pachctl CLI
Console
Create a file named classify.json with the following contents:
{
"pipeline" : {
"name" : "classify" ,
"project" : {
"name" : "standard-ml-pipeline"
},
},
"description" : "Runs the image only model and image+heatmaps model for breast cancer prediction." ,
"input" : {
"cross" : [
{
"join" : [
{
"pfs" : {
"repo" : "crop" ,
"glob" : "/(*)" ,
"joinOn" : "$1"
}
},
{
"pfs" : {
"repo" : "extract_centers" ,
"glob" : "/(*)" ,
"joinOn" : "$1"
}
},
{
"pfs" : {
"repo" : "generate_heatmaps" ,
"glob" : "/(*)" ,
"joinOn" : "$1"
}
}
]
},
{
"pfs" : {
"repo" : "models" ,
"glob" : "/"
}
}
]
},
"transform" : {
"cmd" : [
"/bin/bash" ,
"multi-stage/classify.sh"
] ,
"image" : "pachyderm/breast_cancer_classifier:1.11.6"
},
"resourceLimits" : {
"gpu" : {
"type" : "nvidia.com/gpu" ,
"number" : 1
}
},
"resourceRequests" : {
"memory" : "4G" ,
"cpu" : 1
}
} Copy
Save the file.
Create the pipeline
pachctl create pipeline - f / path / to / classify.json Copy
Select Create > Pipeline .
Overwrite the default json with the following:
{
"pipeline" : {
"name" : "classify" ,
"project" : {
"name" : "standard-ml-pipeline"
},
},
"description" : "Runs the image only model and image+heatmaps model for breast cancer prediction." ,
"input" : {
"cross" : [
{
"join" : [
{
"pfs" : {
"repo" : "crop" ,
"glob" : "/(*)" ,
"joinOn" : "$1"
}
},
{
"pfs" : {
"repo" : "extract_centers" ,
"glob" : "/(*)" ,
"joinOn" : "$1"
}
},
{
"pfs" : {
"repo" : "generate_heatmaps" ,
"glob" : "/(*)" ,
"joinOn" : "$1"
}
}
]
},
{
"pfs" : {
"repo" : "models" ,
"glob" : "/"
}
}
]
},
"transform" : {
"cmd" : [
"/bin/bash" ,
"multi-stage/classify.sh"
] ,
"image" : "pachyderm/breast_cancer_classifier:1.11.6"
},
"resourceLimits" : {
"gpu" : {
"type" : "nvidia.com/gpu" ,
"number" : 1
}
},
"resourceRequests" : {
"memory" : "4G" ,
"cpu" : 1
}
} Copy
4. Upload Dataset
#
Open or download this github repo.
gh repo clone pachyderm / docs - content Copy
Navigate to this tutorial.
cd content / products / mldm / latest / build - dags / tutorials / task - parallelism Copy
Upload the sample_data and models folders to your repos.
Pachctl CLI
Console
pachctl put file - r sample_data @ master - f sample_data /
pachctl put file - r models @ master - f models / Copy
Open the sample_data repo.
Select Upload > Files .
Upload the sample_data folder.
Open the models repo.
Select Upload > Files .
Upload the models folder.
User Code Assets
# The Docker image used in this tutorial was built with the following assets:
Dockerfile
Crop.sh
Extract_Centers.sh
Generate_Heatmaps.sh
Classify.sh
Visualize_Heatmaps.sh
FROM pytorch / pytorch : 1.7.1 - cuda11.0 - cudnn8 - devel
# Update NVIDIA's apt-key
# Announcement: https://forums.developer.nvidia.com/t/notice-cuda-linux-repository-key-rotation/212772
ENV DISTRO ubuntu1804
ENV CPU_ARCH x86_64
RUN apt - key adv -- fetch - keys https :// developer.download.nvidia.com / compute / cuda / repos /$ DISTRO /$ CPU_ARCH / 3 bf863cc.pub
RUN apt - get update && apt - get install - y git libgl1 - mesa - glx libglib2.0 -0
WORKDIR / workspace
RUN git clone https :// github.com / jimmywhitaker / breast_cancer_classifier.git / workspace
RUN pip install -- upgrade pip && pip install - r requirements.txt
RUN pip install matplotlib -- ignore - installed
RUN apt - get - y install tree
COPY . / workspace Copy #!/bin/bash
NUM_PROCESSES = 10
ID =$ ( ls / pfs / sample_data / | head - n 1 )
DATA_FOLDER = "/pfs/sample_data/${ID}/"
INITIAL_EXAM_LIST_PATH = "/pfs/sample_data/${ID}/gen_exam_list_before_cropping.pkl"
CROPPED_IMAGE_PATH = "/pfs/out/${ID}/cropped_images"
CROPPED_EXAM_LIST_PATH = "/pfs/out/${ID}/cropped_images/cropped_exam_list.pkl"
EXAM_LIST_PATH = "/pfs/out/${ID}/data.pkl"
export PYTHONPATH =$ ( pwd ) :$ PYTHONPATH
echo 'Stage 1: Crop Mammograms'
python3 src / cropping / crop_mammogram.py \
-- input - data - folder $ DATA_FOLDER \
-- output - data - folder $ CROPPED_IMAGE_PATH \
-- exam - list - path $ INITIAL_EXAM_LIST_PATH \
-- cropped - exam - list - path $ CROPPED_EXAM_LIST_PATH \
-- num - processes $ NUM_PROCESSES Copy #!/bin/bash
DEVICE_TYPE = 'gpu'
NUM_EPOCHS = 10
HEATMAP_BATCH_SIZE = 100
GPU_NUMBER = 0
ID =$ ( ls / pfs / crop / | head - n 1 )
IMAGEHEATMAPS_MODEL_PATH = "/pfs/models/sample_imageheatmaps_model.p"
CROPPED_IMAGE_PATH = "/pfs/crop/${ID}/cropped_images"
EXAM_LIST_PATH = "/pfs/extract_centers/${ID}/data.pkl"
HEATMAPS_PATH = "/pfs/generate_heatmaps/${ID}/heatmaps"
IMAGEHEATMAPS_PREDICTIONS_PATH = "/pfs/out/${ID}/imageheatmaps_predictions.csv"
export PYTHONPATH =$ ( pwd ) :$ PYTHONPATH
echo 'Stage 4b: Run Classifier (Image+Heatmaps)'
python3 src / modeling / run_model.py \
-- model - path $ IMAGEHEATMAPS_MODEL_PATH \
-- data - path $ EXAM_LIST_PATH \
-- image - path $ CROPPED_IMAGE_PATH \
-- output - path $ IMAGEHEATMAPS_PREDICTIONS_PATH \
-- use - heatmaps \
-- heatmaps - path $ HEATMAPS_PATH \
-- use - augmentation \
-- num - epochs $ NUM_EPOCHS \
-- device - type $ DEVICE_TYPE \
-- gpu - number $ GPU_NUMBER Copy #!/bin/bash
NUM_PROCESSES = 10
ID =$ ( ls / pfs / crop / | head - n 1 )
CROPPED_IMAGE_PATH = "/pfs/crop/${ID}/cropped_images"
CROPPED_EXAM_LIST_PATH = "/pfs/crop/${ID}/cropped_images/cropped_exam_list.pkl"
EXAM_LIST_PATH = "/pfs/out/${ID}/data.pkl"
export PYTHONPATH =$ ( pwd ) :$ PYTHONPATH
echo 'Stage 2: Extract Centers'
python3 src / optimal_centers / get_optimal_centers.py \
-- cropped - exam - list - path $ CROPPED_EXAM_LIST_PATH \
-- data - prefix $ CROPPED_IMAGE_PATH \
-- output - exam - list - path $ EXAM_LIST_PATH \
-- num - processes $ NUM_PROCESSES Copy #!/bin/bash
DEVICE_TYPE = 'gpu'
HEATMAP_BATCH_SIZE = 100
GPU_NUMBER = 0
ID =$ ( ls / pfs / crop / | head - n 1 )
PATCH_MODEL_PATH = "/pfs/models/sample_patch_model.p"
CROPPED_IMAGE_PATH = "/pfs/crop/${ID}/cropped_images"
EXAM_LIST_PATH = "/pfs/extract_centers/${ID}/data.pkl"
HEATMAPS_PATH = "/pfs/out/${ID}/heatmaps"
export PYTHONPATH =$ ( pwd ) :$ PYTHONPATH
echo 'Stage 3: Generate Heatmaps'
python3 src / heatmaps / run_producer.py \
-- model - path $ PATCH_MODEL_PATH \
-- data - path $ EXAM_LIST_PATH \
-- image - path $ CROPPED_IMAGE_PATH \
-- batch - size $ HEATMAP_BATCH_SIZE \
-- output - heatmap - path $ HEATMAPS_PATH \
-- device - type $ DEVICE_TYPE \
-- gpu - number $ GPU_NUMBER Copy #!/bin/bash
ID =$ ( ls / pfs / crop / | head - n 1 )
CROPPED_IMAGE_PATH = "/pfs/crop/${ID}/cropped_images/"
HEATMAPS_PATH = "/pfs/generate_heatmaps/${ID}/heatmaps/"
OUTPUT_PATH = "/pfs/out/${ID}/"
export PYTHONPATH =$ ( pwd ) :$ PYTHONPATH
echo 'Stage 5: Visualize Heatmaps'
python3 src / heatmaps / visualize_heatmaps.py \
-- image - path $ CROPPED_IMAGE_PATH \
-- heatmap - path $ HEATMAPS_PATH \
-- output - path $ OUTPUT_PATH Copy